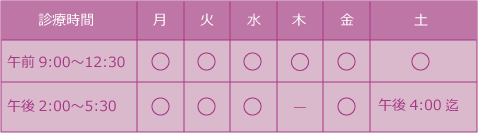
HLA-DRについて
ヒト白血球型抗原(ヒトはっけっきゅうがたこうげん、Human Leukocyte Antigen; HLA)とはヒトの主要組織適合遺伝子複合体のことである。白血球の血液型と言えるものであり、一般的に血液型というとA、O、AB、B型といった赤血球の型を指すが、HLA型は白血球の型を示している。ただし、白血球以外にもHLAは存在するため、現在ではヒト白血球型抗原の名称で呼ばれることはほとんどなく、HLAと略して呼ばれる。
概要
ヒト白血球型抗原は、第6染色体短腕上に存在する主要組織適合遺伝子複合体(MHC)の産物である。その型の種類は多く、まずA座のA1、A2、A210(2)、A3…A80、B座のB5、B7、B703(7)…、C座の…、DR座の…と続き赤血球の型とは比較にならないほど膨大で、その組み合わせは数万通りといわれる。
クラスI抗原の各α鎖遺伝子は主に6種の遺伝子(A~G)からなり、α1とα2ドメインをコードする遺伝子の第2エクソンと第3エクソン内に多型性を示す領域がある。
クラスII抗原のDR抗原は複数の遺伝子から構成され、DRB1遺伝子がコードするβ鎖とDRA遺伝子がコードするα鎖のヘテロダイマーにより、血清学的特異性のDR1~18に対応する抗原が構成される。また、DRB3、DRB4、DRB5遺伝子がコードするβ鎖とDRA遺伝子がコードするα鎖のヘテロダイマーにより、DR52、DR53、DR51に対応する抗原が構成される。
同様にDQ抗原もDQB1遺伝子がコードするβ鎖とDRA遺伝子がコードするα鎖のヘテロダイマーにより、DQ1~DQ9に対応する抗原が構成される。
またDP抗原もDPB1遺伝子がコードするβ鎖とDRA遺伝子がコードするα鎖のヘテロダイマーにより、DPw1~DPw6に対応する抗原が構成される。
クラスII遺伝子の中で抗原ペプチド結合部に多形型を示すのはDRB1、DRB3、DRB4、DRB5、DQA1、DQB1、DPA1、DPB1であり、それぞれのβ1とα2ドメインをコードする遺伝子の第2エクソン内に塩基置換がみられる。
献血の際に献血者が登録(献血者登録制)することにより、HLA適合血小板を必要としている患者に対して、HLAが適合した輸血を行なうことができる。
クラスI抗原
クラスIa抗原(HLA-A、HLA-B、HLA-C)、クラスIb抗原(HLA-E、HLA-F、HLA-G)に分けられ、ほとんどの有核細胞や血小板、血漿中にある。
前者はキラーT細胞の誘導において拘束分子として機能する。HLA-A3、HLA-A11、HLA-Bw4、HLA-CがNK細胞レセプターのリガンドであり、NK活性を抑制する。
ウイルス感染細胞や癌細胞は免疫応答から逃れるために、自身のクラスI抗原を消失させるが、逆にNK細胞はHLA抗原を失った細胞によって活性化される。
HLA-Gだけは胎盤トロホブラストに特異的に存在し(胎盤トロホブラストには他のクラスI、IIは無い)胎児保護のために母体のNK活性やキラーT細胞の抑制をしている。
また赤血球抗原のBga、Bgb、Bgc抗原はクラスI抗原のHLA-B7、B17、A28抗原と同じである。
クラスI抗原のL鎖はβ2-ミクログロブリンである。
クラスII抗原
マクロファージや単球等の抗原提示細胞やBリンパ球、活性化Tリンパ球などに分布している。HLA-DR、HLA-DQ、HLA-DP抗原はそれぞれの遺伝子によってコードされ、ヘルパーT細胞やサプレッサーT細胞の誘導に拘束分子として機能する。
クラスII抗原はα鎖β鎖ヘテロダイマーに関係があり、さらにCD4分子と親和性がある。
クラスIII抗原
補体成分やTNF(腫瘍壊死因子)などを支配している。
HLA抗原と相関がある疾患は、インスリン依存性糖尿病やぶどう膜炎、先天性副腎皮質過形成症やベーチェット病、強直性脊椎炎など多数ある。
日本人の成り立ち
HLAハプロタイプは日本人の成り立ちに重要な示唆を与える。徳永勝士によると、日本人には大きく以下の4タイプの流れが認められる[1][2][3][4]。
- B52-DR2: 中国大陸北部から朝鮮半島を経て北九州・近畿へ
- B44-DR13、B7-DR1: 満州・朝鮮半島東部から日本海沿岸へ
- B54-DR4: 中国南部から琉球諸島を経て太平洋側へ
- B46-DR8: 中国大陸南部から直接、あるいは朝鮮半島を経由して北九州へ
1.は中国北部、モンゴルの一集団に高頻度のタイプで、国内では九州北部から本州中央部にかけて多い。2.は満州族、朝鮮民族に高頻度タイプで、国内では日本海側に多い。3.は中国南部に多いタイプで、国内では沖縄や太平洋側に多い。4.は国外では満州族と朝鮮民族のみにみられ、国内には九州北部から本州中央部にかけて多い。このタイプの姉妹タイプB46-DR9が東南アジアで最も高頻度でみられる。
さらにこれとは別に縄文系と想定される別の複数のハプロタイプが南九州や北東北に存在する。またアイヌは日本人と異なる型が多いという。
HLA-DR
| MHC class II, DR | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (heterodimer) | |||||||||||||||||||
Illustration of DR with bound ligand (yellow) | |||||||||||||||||||
| Protein type | cell surface receptor | ||||||||||||||||||
| Function | Immune recognition and antigen presentation | ||||||||||||||||||
| |||||||||||||||||||
HLA-DR is an MHC class IIcell surface receptor encoded by the human leukocyte antigen complex on chromosome 6 region 6p21.31. The complex of HLA-DR (Human Leukocyte Antigen – DR isotype) and peptide, generally between 9 and 30 amino acids in length, constitutes a ligand for the T-cell receptor (TCR). HLA (human leukocyte antigens) were originally defined as cell surface antigens that mediate graft-versus-host disease. Identification of these antigens has led to greater success and longevity in organ transplant.
Antigens most responsible for graft loss are HLA-DR (first six months), HLA-B (first two years), and HLA-A (long-term survival).[1] Good matching of these antigens between host and donor is most critical for achieving graft survival.
HLA-DR is also involved in several autoimmune conditions, disease susceptibility and disease resistance. It is also closely linked to HLA-DQ and this linkage often makes it difficult to resolve the more causative factor in disease.
HLA-DR molecules are upregulated in response to signalling. In the instance of an infection, the peptide (such as the staphylococcal enterotoxin I peptide) is bound into a DR molecule and presented to a few of a great many T-cell receptors found on T-helper cells. These cells then bind to antigens on the surface of B-cells stimulating B-cell proliferation.
Function

The primary function of HLA-DR is to present peptide antigens, potentially foreign in origin, to the immune system for the purpose of eliciting or suppressing T-(helper)-cell responses that eventually lead to the production of antibodies against the same peptide antigen. Antigen-presenting cells (macrophages, B-cells and dendritic cells) are the cells in which DR are typically found. Increased abundance of DR 'antigen' on the cell surface is often in response to stimulation, and, therefore, DR is also a marker for immune stimulation.
Structure
HLA-DR is an αβ heterodimer, cell surface receptor, each subunit of which contains two extracellular domains, a membrane-spanning domain and a cytoplasmic tail. Both α and β chains are anchored in the membrane. The N-terminal domain of the mature protein forms an alpha-helix that constitutes the exposed part of the binding groove, the C-terminal cytoplasmic region interact with the other chain forming a beta-sheet under the binding groove spanning to the cell membrane. The majority of the peptide contact positions are in the first 80 residues of each chain.
Genetics
The genetics of HLA-DR is complex. HLA-DR is encoded by several loci and several 'genes' of different function at each locus. The DR α-chain is encoded by the HLA-DRAlocus. Unlike the other DR loci, functional variation in mature DRA gene products is absent. (Note: see table Number of Variant Alleles HLA-DR Loci) This reduces the potential functional combinations from ~1400 to ~400 ([table is not exact because new alleles are continually being added; not all new alleles are functional variants of the mature subunits]).
| DR | DR-DQ | DR | DQ | Freq | ||
|---|---|---|---|---|---|---|
| Serotype | haplotype | B1 | A1 | B1 | %[2] | |
| DR1 | DR1-DQ5 | 01:01 | 01:01 | 05:01 | 9. | 1 |
| 01:02 | 01:01 | 05:01 | 1. | 4 | ||
| 01:03 | 01:01 | 05:01 | 0. | 5 | ||
| DR3 | DR3-DQ2 | 03:01 | 05:01 | 02:01 | 13. | 1 |
| DR4 | DR4-DQ7 | 04:01 | 0300 | 03:01 | 5. | 4 |
| 04:07 | 0300 | 03:01 | 0. | 9 | ||
| DR4-DQ8 | 04:01 | 0300 | 03:02 | 5. | 0 | |
| 04:02 | 0300 | 03:02 | 1. | 0 | ||
| 04:03 | 0300 | 03:02 | 0. | 4 | ||
| 04:04 | 0300 | 03:02 | 3. | 9 | ||
| 04:05 | 0300 | 03:02 | 0. | 3 | ||
| DR7 | DR7-DQ2 | 07:01 | 02:01 | 02:02 | 11. | 1 |
| DR7-DQ9 | 07:01 | 02:01 | 03:03 | 3. | 7 | |
| DR8 | DR8-DQ4 | 08:01 | 04:01 | 04:02 | 2. | 2 |
| DR8-DQ7 | 08:03 | 06:01 | 03:01 | 0. | 1 | |
| DR9 | DR9-DQ9 | 09:01 | 0300 | 03:03 | 0. | 8 |
| DR10 | DR10-DQ5 | 10:01 | 01:04 | 05:01 | 0. | 7 |
| DR11 | DR11-DQ7 | 11:01 | 05:05 | 03:01 | 5. | 6 |
| 11:03 | 05:05 | 03:01 | 0. | 3 | ||
| 11:04 | 05:05 | 03:01 | 2. | 7 | ||
| DR12 | DR12-DQ7 | 12:01 | 05:05 | 03:01 | 1. | 1 |
| DR13 | DR13-DQ6 | 13:01 | 01:03 | 06:03 | 5. | 6 |
| 13:02 | 01:02 | 06:04 | 3. | 4 | ||
| 13:02 | 01:02 | 06:09 | 0. | 7 | ||
| DR13-DQ7 | 13:03 | 05:05 | 03:01 | 0. | 7 | |
| DR14 | DR14-DQ5 | 14:01 | 01:04 | 05:03 | 2. | 0 |
| DR15 | DR15-DQ6 | 15:01 | 01:02 | 06:02 | 14. | 2 |
| 15:02 | 01:03 | 06:01 | 0. | 7 | ||
| DR16 | DR16-DQ5 | 16:01 | 01:02 | 05:02 | 1. | 0 |
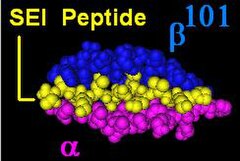
The DR β-chain[3] is encoded by 4 loci, however no more than 3 functional loci are present in a single individual, and no more than two on a single chromosome. Sometimes an individual may only possess 2 copies of the same locus, DRB1*. The HLA-DRB1 locus is ubiquitous and encodes a very large number of functionally variable gene products (HLA-DR1 to HLA-DR17). The HLA-DRB3 locus encodes the HLA-DR52 specificity, is moderately variable and is variably associated with certain HLA-DRB1 types. The HLA-DRB4 locus encodes the HLA-DR53 specificity, has some variation, and is associated with certain HLA-DRB1 types. The HLA-DRB5 locus encodes the HLA-DR51 specificity, which is typically invariable, and is linked to the HLA-DR2 types.
- linkage (See Table)
- DQA1 and DQB1
- Linkage disequilibrium exists for many DR-DQ types.
- Nomenclature issues. Some older studies may refer to DR15 or 16 as DR2 and DQ5 and DQ6 as DQ1 therefore a haplotype DR2-DQ1 is usually referring to DR15-DQ6 but could be referring to DR16-DQ5. DR5 is used to refer to DR11 and DR12, in which case DQ3 might be used. In these instances DQ3 almost always can be interpreted as DQ7, but DR5 is most often DR11 and less frequently DR12. Similar issues exist for DR6 versus DR13 and DR14. DR6-DQ1 can refer to either DR13-DQ6 or less frequently DR14-DQ5, but DR6-DQ3 or DR6-DQ7 generally refers to DR13-DQ7. Even older literature has more confusing designations. By looking at the change of disease association with improved testing we can see how HLA nomenclature has evolved over time.
- DQA1 and DQB1
| HLA-DR | ||||
|---|---|---|---|---|
| HLA | -A1 | -B1 | -B3 to -B51 | Potential |
| Locus | # | # | # | Combinations |
| Alleles[3][4] | 3 | 463 | 74 | 1635 |
| Unique Polypeptide | 2 | 394 | 57 | 902 |
| Contact Variant | 1 | ~300 | ~30 | ~330 |
| 1DRB3, DRB4, DRB5 have variable presence in humans | ||||
Evolution and allele frequencies
[edit]There is a high level of allelic diversity at HLA DRB1, it is second only to HLA-B locus in number of allelic variants. These two loci are highest sequence variation rate within human genome. This means HLA-DRB1 is rapidly evolving, much more rapidly than almost all other protein encoding loci. Much of the variation at HLA DRB1 occurs at peptide contact positions in the binding groove, as a result many of the alleles alter the way the DR binds peptide ligands and changes the repertoire each receptor can bind. This means that most of the changes are functional in nature, and therefore are under selection. In the HLA region, genes are under heterozygous or balancing selection, although certain alleles appear to be under positive or negative selection, either in the past or present
HLA generally evolve through a process of gene conversion, which is a form of short distance or 'abortive' genetic recombination. Functional motifs in genes are exchanged to form new alleles, and frequently new, functionally different DR isoforms. HLA-DR represents an extreme example of this. A survey of X-linked loci reveals that most human loci have undergone fixation within the last 600,000 years, and diploid loci have undergone significant proportion of fixation in that period of time.
The level of deep branching at X-linked loci indicates loci were close to fixation or fixed at the end of the human population bottleneck 100,000 to 150,000 years ago. The HLA-DR locus represents a major exception to this observation.[5] Based on distribution of major groupings in the human population it is possible to assert that more than a dozen major variants survived the population bottleneck. This observation is supported by the concept of a heterozygous selection coefficient operating on the HLA-DR, and at the HLA-DRB1 locus to a greater degree relative to HLA-DQB1 and HLA-DPB1. Most of the HLA alleles currently present in the human population can be explained by gene conversion between these ancient ancestral types,[6] some that persist into the extant population.
Serogroups
| Serotypes of HLA-DRB1 gene products | ||
| Split antigens | ||
| HLA-DR1 | ||
| HLA-DR2 | HLA-DR15 | HLA-DR16 |
| HLA-DR3 | HLA-DR17 | HLA-DR18 |
| HLA-DR4 | ||
| HLA-DR5 | HLA-DR11 | HLA-DR12 |
| HLA-DR6 | HLA-DR13 | HLA-DR14 |
| HLA-DR7 | ||
| HLA-DR8 | ||
| HLA-DR9 | ||
| HLA-DR10 | ||
The table below provides links to subpages with information about distribution, genetic linkage and disease association for the HLA-DR serogroups.
Interlocus DRB linkage
DRB1 is linked with other DRB loci in four ways.
| non-DRB1 | linked DRB1 antigens | |||
|---|---|---|---|---|
| antigens | antigens | |||
| None | DR1 | DR8 | DR10 | |
| DR51 | DR2 | DR15 | DR16 | |
| DR52 | DR3 | DR17 | DR18 | |
| DR5 | DR11 | DR12 | ||
| DR6 | DR13 | DR14 | ||
| DR53 | DR4 | DR7 | DR8 | DR9 |
2025年5月11日 | カテゴリー:免疫疾患 |